Topic: 3D Imaging and quantitative analysis of intact tissues and organs
My PhD thesis focuses on imaging methods that enable visualization and analysis of tissue and organs in 3D over a period of time, which leads to a better understating of tissue development and maintenance.
Organs and tissues are highly complex structures that come together to form whole organisms. Scientists have spent hundreds of years fully examining elements of the human body, yet little is known about its complexity. The main reason is that organs and tissues are highly heterogeneous. Each organ has a three-dimensional (3D) architecture, which is composed of various cells and environments that may change over time. This is why the features of organs/tissues need to be investigated in space and time to fully comprehend their organization and functions. If we look at an organ, we can see its gross anatomy, but we cannot see its cellular and molecular composition as well as dynamic changes. Although the human body is mostly made of water, we do not see what is deep inside organs as they are not transparent. The reason is that each cell is composed of different molecules, such as proteins, lipids and cytosol. Their unique properties cause bending (diffracting) of light when it is passing through an organ. As our body is made of billions of various cells, the light is highly diffracted, which causes tissues to be opaque. Additionally, most cells are too small to see with the naked eye. Thus, imaging is a great tool to obtain detailed information with spatiotemporal resolution. Although there are many well-established methods of imaging, challenges persist.
The standard procedure for tissue imaging is first to cut a sample into small slices to see structures deep inside it. Then, elements of cells (proteins and RNAs) are labeled with a fluorescent tag and visualized under a microscope. These tags absorb the light and emit a visible signal. However, most microscopes have a limited imaging depth, meaning that only several dozen micrometers of a tissue surface can be visualized. Thus, as mentioned above, the standard procedure involves two-dimensional (2D) slices of tissues. Cutting even a small piece of an organ is very labor- and time-intensive work. Moreover, 2D slices do not represent real volumetric architecture. Understanding an organ/tissue in 3D is crucial, for example, as pathologists who diagnose cancer patients may miss some determining features if they investigate only a few thin slices of a tumor biopsy. Moreover, standard 2D procedures involve slices, which are not viable. Understanding the cellular dynamics that shape organs/tissues requires imaging living cells over a period of time.
The first aim of the thesis was to visualize a whole tissue without actually cutting it into smaller pieces. The second aim was to develop a method to label not only proteins but also RNA in large samples, which has not yet been achievable. After labeling both proteins and RNAs, we made the tissues transparent by ‘clearing’ them with a set of chemicals reduce the diffraction of the light. Next, we chose a microscope that was fast and efficient to scan a few centimeters of tissue in a few hours. As a last step, we stored a large number acquired of images, visualized the results and performed quantitative analysis. We have developed this method for clinical human tumor biopsies that are stored in a very established and specific way. Human tumors are highly heterogeneous tissues with various populations of cells existing in specific areas. These distinct populations as well as their environmental features, such as blood vessel compositions, can only be properly examined in 3D. Thus, by using algorithms and special software, we discovered new tumor characteristics. In comparison to standard 2D strategies, our new methods improved the characterization and staging of cancer and consequently may lead to more accurate diagnosis and treatment strategies for patients.
The last aim was to examine tissue dynamics over time. Since it is estimated that we are limited to understanding only a small percentage of brain functionality, we used imaging techniques to investigate brain development. As cells needed to be viable for this analysis, we used living tissues dissected from mice and cell cultures in a dish. We have found that certain proteins contribute to spontaneous calcium signaling, which is one of the most important ways for cells to communicate with each other. This process stimulates the creation of new neurons in the developing brain.
To summarize, the imaging in space and time performed in the thesis led to a better understanding of physiological and pathological development.
#themanyfacesofscience #scienceforall
Source: https://openarchive.ki.se/xmlui/handle/10616/47109
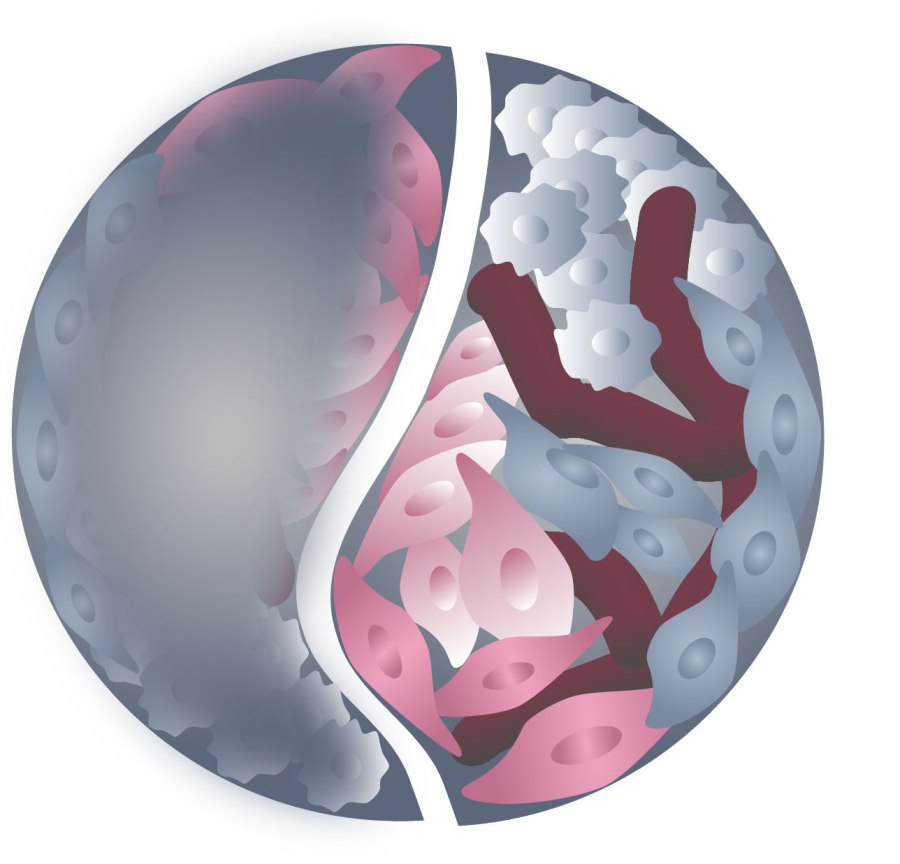
Illustration represents a 3D heterogeneous tissue. The left blurry part represents a tissue without tissue-clearing methods and the right part shows a tissue after clearing by Dagmara Kaczynska.
Featured image: Gordon Johnson from Pixabay

0 comments